HugeCTR Python Interface
About the HugeCTR Python Interface
As a recommendation system domain specific framework, HugeCTR has a set of high level abstracted Python Interface which includes training API and inference API. Users only need to focus on algorithm design, the training and inference jobs can be automatically deployed on the specific hardware topology in the optimized manner. From version 3.1, users can complete the process of training and inference without manually writing JSON configuration files. All supported functionalities have been wrapped into high-level Python APIs. Meanwhile, the low-level training API is maintained for users who want to have precise control of each training iteration and each evaluation step. Still, the high-level training API is friendly to users who are already familiar with other deep learning frameworks like Keras and it is worthwhile to switch to it from low-level training API. Please refer to HugeCTR Python Interface Notebook to get familiar with the workflow of HugeCTR training and inference. Meanwhile we have a lot of samples for demonstration in the samples directory of the HugeCTR repository.
High-level Training API
For HugeCTR high-level training API, the core data structures are Solver, EmbeddingTrainingCacheParams, DataReaderParams, OptParamsPy, Input, SparseEmbedding, DenseLayer and Model. You can create a Model instance with Solver, EmbeddingTrainingCacheParams, DataReaderParams and OptParamsPy instances, and then add instances of Input, SparseEmbedding or DenseLayer to it. After compiling the model with the Model.compile() method, you can start the epoch mode or non-epoch mode training by simply calling the Model.fit() method. Moreover, the Model.summary() method gives you an overview of the model structure. We also provide some other methods, such as saving the model graph to a JSON file, constructing the model graph based on the saved JSON file, loading model weights and optimizer status, etc.
Solver
CreateSolver method
hugectr.CreateSolver()
CreateSolver returns an Solver object according to the custom argument values,which specify the training resources.
Arguments
model_name: String, the name of the model. The default value is empty string. If you want to dump the model graph and save the model weights for inference, a unique value should be specified for each model that needs to be deployed.seed: A random seed to be specified. The default value is 0.lr_policy: The learning rate policy which suppots only fixed. The default value isLrPolicy_t.fixed.lr: The learning rate, which is also the base learning rate for the learning rate scheduler. The default value is 0.001.warmup_steps: The warmup steps for the internal learning rate scheduler within Model instance. The default value is 1.decay_start: The step at which the learning rate decay starts for the internal learning rate scheduler within Model instance. The default value is 0.decay_steps: The number of steps of the learning rate decay for the internal learning rate scheduler within Model instance. The default value is 1.decay_power: The power of the learning rate decay for the internal learning rate scheduler within Model instance. The default value is 2.end_lr: The final learning rate for the internal learning rate scheduler within Model instance. The default value is 0. Please refer to SGD Optimizer and Learning Rate Scheduling if you want to get detailed information about LearningRateScheduler.max_eval_batches: Maximum number of batches used in evaluation. It is recommended that the number is equal to or bigger than the actual number of bathces in the evaluation dataset. The default value is 100.batchsize_eval: Minibatch size used in evaluation. The default value is 2048. Note that batchsize here is the global batch size across gpus and nodes, not per worker batch size.batchsize: Minibatch size used in training. The default value is 2048. Note that batchsize here is the global batch size across gpus and nodes , not per worker batch size.vvgpu: GPU indices used in the training process, which has two levels. For example: [[0,1],[1,2]] indicates that two physical nodes (each physical node can have multiple NUMA nodes) are used. In the first node, GPUs 0 and 1 are used while GPUs 1 and 2 are used for the second node. It is also possible to specify non-continuous GPU indices such as [0, 2, 4, 7]. The default value is [[0]].repeat_dataset: Whether to repeat the dataset for training. If the value isTrue, non-epoch mode training will be employed. Otherwise, epoch mode training will be adopted. The default value isTrue.use_mixed_precision: Whether to enable mixed precision training. The default value isFalse.enable_tf32_compute: If you want to accelerate FP32 matrix multiplications within the FullyConnectedLayer and InteractionLayer, set this value toTrue. The default value isFalse.scaler: The scaler to be used when mixed precision training is enabled. Only 128, 256, 512, and 1024 scalers are supported for mixed precision training. The default value is 1.0, which corresponds to no mixed precision training.metrics_spec: Map of enabled evaluation metrics. You can use either AUC, AverageLoss, HitRate, or any combination of them. For AUC, you can set its threshold, such as {MetricsType.AUC: 0.8025}, so that the training terminates when it reaches that threshold. The default value is {MetricsType.AUC: 1.0}. Multiple metrics can be specified in one job. For example: metrics_spec = {hugectr.MetricsType.HitRate: 0.8, hugectr.MetricsType.AverageLoss:0.0, hugectr.MetricsType.AUC: 1.0})i64_input_key: If your dataset format isNorm, you can choose the data type of each input key. For theParquetformat dataset generated by NVTabular, only I64 is allowed. For theRawdataset format, only I32 is allowed. Set this value toTruewhen you need to use I64 input key. The default value isFalse.use_algorithm_search: Whether to use algorithm search for cublasGemmEx within the FullyConnectedLayer. The default value isTrue.use_cuda_graph: Whether to enable cuda graph for dense network forward and backward propagation. The default value isTrue.device_layout: this option is deprecated and no longer used.use_holistic_cuda_graph: The default value isFalse. IfTrue, everything inside a training iteration is packed into a CUDA Graph. Requirement: use_cuda_graph is False, use_overlapped_pipeline is True.use_overlapped_pipeline: The default value isFalse. IfTrue, the computation in the dense input data path will be overlapped with the hybrid embedding computation. Requirements: The data reader is asynchronous (see AsyncParam), hybrid embedding is used (see HybridEmbeddingParam), the model has a feature interaction layer (see InteractionLayer).data_source_params: DataSourceParams(), specify the configurations of the data sources(Local, HDFS, or others) for model persistence and loading.all_reduce_algo: The algorithm to be used for all reduce. The supported options areAllReduceAlgo.OneShotandAllReduceAlgo.NCCL. The default value isAllReduceAlgo.NCCL. When you are doing multi-node training,AllReduceAlgo.OneShotwill require RDMA support whileAllReduceAlgo.NCCLcan run on both RDMA and non-RDMA hardware.grouped_all_reduce: The default value isFalse. IfTrue, the gradients for the dense network and the gradients for data-parallel embedding are grouped and all reduced in one kernel, effectively combining two small all-reduce operations into a single larger one for higher efficiency. Requirements: Hybrid embedding is used (see HybridEmbeddingParam).num_iterations_statistics: The number of batches used to perform statistics for hybrid embedding. The default value is20. Requirement: The data reader is asynchronous (see AsyncParam).
Example:
solver = hugectr.CreateSolver(max_eval_batches = 300,
batchsize_eval = 16384,
batchsize = 16384,
lr = 0.001,
vvgpu = [[0]],
repeat_dataset = True,
i64_input_key = True)
CreateETC method
hugectr.CreateETC()
CreateETC should only be called when using the Embedding Training Cache (ETC) feature. It returns a EmbeddingTrainingCacheParams object that specifies the parameters for initializing a EmbeddingTrainingCache instance.
Arguments
ps_types: A list specifies types of parameter servers (PS) of each embedding table. Available PS choices for embeddings are:-
The whole embedding table will be loaded into the host memory in the initialization stage.
It requires the size of host memory should be large enough to hold the embedding table along with the optimizer states (if any).
Stagedtype offers better loading and dumping bandwidth than theCachedPS.
-
A sub-portion of the embedding table will be dynamically cached in the host memory, and it adopts a runtime eviction/insertion mechanism to update the cached table.
The size of the cached table is configurable, which can be substantially smaller than the size of the embedding table stored in the SSD or various kinds of filesystems. E.g., embedding table size (1 TB) v.s. cache size (100 GB).
The bandwidth of
CachedPS is mainly affected by the hit rate. If the hit rate is 100 %, its bandwidth tends to theStagedPS; Otherwise, if the hit rate is 0 %, the bandwidth equals the random-accessing bandwidth of SSD.
-
sparse_models: A path list of embedding table(s). If the provided path points to an existing table, this table will be used for incremental training. Otherwise, the newly generated table will be written into this path after training.local_paths: A path list for storing the temporary embedding table. Its length should be equal to the number of MPI ranks. Each entry in this list should be a path pointing to the local SSD of this node.This entry is only required when there is
hugectr.TrainPSType_t.Cachedinps_types.hcache_configs: A path list of the configurations ofCachedPS. Please check Cached-PS Configuration for more descriptions.If only one configuration is provided, it will be used for all
CachedPS.Otherwise, you need to provide one configuration for each
CachedPS. And the ith configuration inhcache_configswill be used for the ith occurrence ofCachedPS inps_types.
This entry is only required when there is
hugectr.TrainPSType_t.Cachedinps_types.
Note that the Staged and Cached PS can be used together for a model with more than one embedding tables.
Example usage of the CreateETC() API can be found in Configuration.
For the usage of the ETC feature in real cases, please check the HugeCTR Continuous Training notebook.
AsyncParam
AsyncParam class
hugectr.AsyncParam()
A data reader can be optimized using asynchronous reading. This is done by creating the data reader with a async_param argument (see DataReaderParams), which is of type AsyncParam. AsyncParam specifies the parameters related to asynchronous raw data reader, An asynchronous data reader uses the Linux asynchronous I/O library (AIO) to achieve peak I/O throughput. Requirements: The input dataset has only one-hot feature items and is in raw format.
Arguments
num_threads: Integer, the number of the data reading threads, should be at least 1 per GPU。 There is NO default value.num_batches_per_thread: Integer, the number of the batches each data reader thread works on simultaneously, typically 2-4. There is NO default value.io_block_size: Integer, the size of individual IO requests, the value 512000 should work in most cases. There is NO default value.io_depth: Integer, the size of the asynchronous IO queue, the value 4 should work in most cases. There is NO default value.io_alignment: Integer, the byte alignment of IO requests, the value 512 should work in most cases. There is NO default value.shuffle: Boolean, if this option is enabled, the order in which the batches are fed into training will be randomized. There is NO default value.aligned_type: The supported types includehugectr.Alignment_t.Autoandhugectr.Alignment_t.Non. Ifhugectr.Alignment_t.Autois chosen, the dimension of dense input will be padded to an 8-aligned value. There is NO default value.
Example:
async_param = hugectr.AsyncParam(32, 4, 716800, 2, 512, True, hugectr.Alignment_t.Non)
HybridEmbeddingParam
HybridEmbeddingParam class
hugectr.HybridEmbeddingParam()
A sparse embedding layer can be optimized using hybrid embedding. Hybrid embedding is designed to overcome the bandwidth constraint that is imposed by the embedding part of the embedding training workload by algorithmically reducing the traffic over the network. Hybrid embedding can improve performance in multi-node and multi-GPU deployments with one-hot data. Conversely, hybrid embedding does not improve performance on a single-machine and single-GPU deployment or with multi-hot encoded data.
You can use hybrid embedding by creating a sparse embedding layer with a hybrid_embedding_param argument that is of type HybridEmbeddingParam and specifying the parameters that are related to hybrid embedding.
Requirements: The input dataset has only one-hot feature items and the model uses the SGD optimizer.
For information about creating a sparse embedding layer, refer to the class documentation.
Arguments
max_num_frequent_categories: Integer, the maximum number of frequent categories in unit of batch size. This argument does not have a default value.max_num_infrequent_samples: Integer, the maximum number of infrequent samples in unit of batch size. This argument does not have a default value.p_dup_max: Float, the maximum probability that the category appears more than once within the gpu-batch. This way of determining the number of frequent categories is used in single-node or NVLink connected systems only. This argument does not have a default value.max_all_reduce_bandwidth: Float, the algorithmic bandwidth of the all reduce. This argument does not have a default value.max_all_to_all_bandwidth: Float, the algorithmic bandwidth of the all-to-all. The unit of bandwidth is per-GPU. This argument does not have a default value.efficiency_bandwidth_ratio: Float, this argument is used in combination withmax_all_reduce_bandwidthandmax_all_to_all_bandwidthto determine the optimal threshold for the number of frequent categories. This way of determining the frequent categories is used for multi node only. This argument does not have a default value.communication_type: The type of communication that is being used. The supported types includeCommunicationType.IB_NVLink,CommunicationType.IB_NVLink_HierandCommunicationType.NVLink_SingleNode. This argument does not have a default value.hybrid_embedding_type: The type of hybrid embedding, which supports onlyHybridEmbeddingType.Distributedfor now. This argument does not have a default value.
Example:
hybrid_embedding_param = hugectr.HybridEmbeddingParam(2, -1, 0.01, 1.3e11, 1.9e11, 1.0,
hugectr.CommunicationType.IB_NVLink_Hier,
hugectr.HybridEmbeddingType.Distributed))
DataReaderParams
DataReaderParams class
hugectr.DataReaderParams()
DataReaderParams specifies the parameters related to the data reader. HugeCTR currently supports three dataset formats, i.e., Norm, Raw and Parquet. An DataReaderParams instance is required to initialize the Model instance.
Arguments
data_reader_type: The type of the data reader which should be consistent with the dataset format. Specify one of the following types:hugectr.DataReaderType_t.Normhugectr.DataReaderType_t.Rawhugectr.DataReaderType_t.ParquetDataReaderType_t.RawAsync
source: List[str] or String, the training dataset source. For Norm or Parquet dataset, specify the file list of training data, such assource = "file_list.txt". For Raw dataset, specify a single training file, such assource = "train_data.bin". When using the embedding training cache, you can specify several file lists, such assource = ["file_list.1.txt", "file_list.2.txt"]. This argument has no default value and you must specify a value.keyset: List[str] or String, the keyset files. This argument is only valid when you use the embedding training cache. The value should correspond to the value for thesourceargument. For example, you can specifysource = ["file_list.1.txt", "file_list.2.txt"]andkeyset = ["file_list.1.keyset", "file_list.2.keyset"]The example shows the one-to-one correspondence between thesourceandkeysetvalues.eval_source: String, the evaluation dataset source. For Norm or Parquet dataset, specify the file list of the evaluation data. For Raw dataset, specify a single evaluation file. This argument has no default value and you must specify a value.check_type: The data error detection mechanism. Specifyhugectr.Check_t.Sum(CheckSum) orhugectr.Check_t.Non(no detection). This argument has no default value and you must specify a value.cache_eval_data: Integer, the cache size of evaluation data on device. Specify a value that is greater than zero to restrict the memory use. The default value is 0.num_samples: Integer, the number of samples in the training dataset. This argument is valid for the Raw dataset format only. The default value is 0.eval_num_samples: Integer, the number of samples in the evaluation dataset. This argument is valid for the Raw dataset format only. The default value is 0.float_label_dense: Boolean, this argument is valid for the Raw dataset format only. When set toTrue, the label and dense features for each sample are interpreted as float values. Otherwise, they are read as integer values while the dense features are preprocessed with \(log(dense[i] + \text{1.f})\). The default value isTrue.num_workers: Integer, the number of data reader workers to load data concurrently. You can empirically decide the best value based on your dataset and training environment. The default value is 12.slot_size_array: List[int], specify the maximum key value for each slot. Refer to the following equation. The array should be consistent with that of the sparse input. HugeCTR requires this argument for Parquet format data and RawAsync format when you want to add an offset to the input key. The default value is an empty list.The following equation shows how to determine the values to specify:
\(slot\_size\_array[k] = \max\limits_i slot^k_i + 1\)
data_source_params: DataSourceParams(), specify the configurations of the data sources(Local, HDFS, or others) for data reading.async_param: AsyncParam, the parameters for async raw data reader. Please find more information in theAsyncParamsection in this document.
Dataset formats
We support the following dataset formats within our DataReaderParams.
Norm
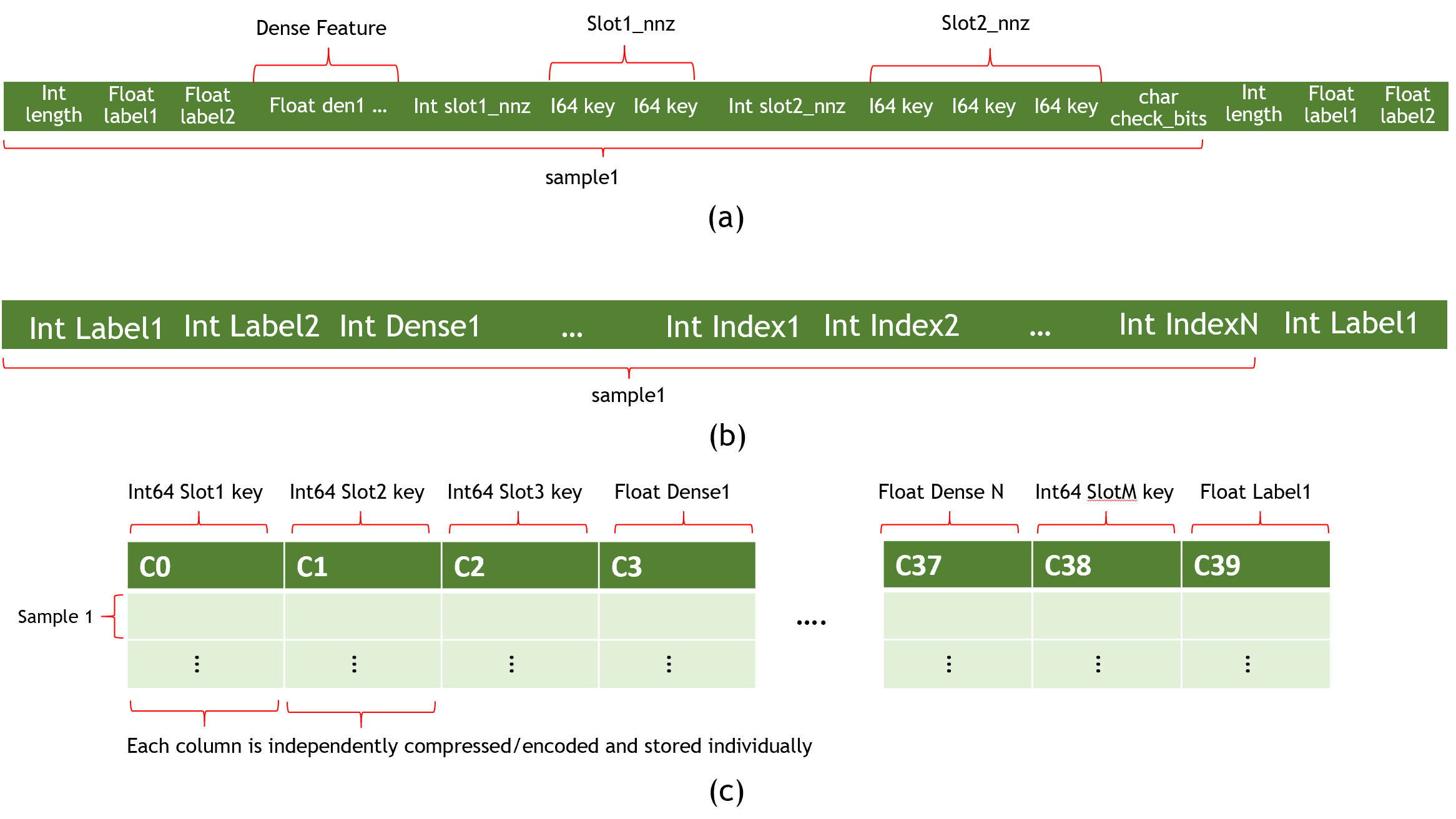
To maximize the data loading performance and minimize the storage, the Norm dataset format consists of a collection of binary data files and an ASCII formatted file list. The model file should specify the file name of the training and testing (evaluation) set, maximum elements (key) in a sample, and the label dimensions as shown in Fig. 1 (a).
Data Files
A data file is the minimum reading granularity for a reading thread, so at least 10 files in each file list are required to achieve the best performance. A data file consists of a header and actual tabular data.
Header Definition:
typedef struct DataSetHeader_ {
long long error_check; // 0: no error check; 1: check_num
long long number_of_records; // the number of samples in this data file
long long label_dim; // dimension of label
long long dense_dim; // dimension of dense feature
long long slot_num; // slot_num for each embedding
long long reserved[3]; // reserved for future use
} DataSetHeader;
Data Definition (each sample):
typedef struct Data_ {
int length; // bytes in this sample (optional: only in check_sum mode )
float label[label_dim];
float dense[dense_dim];
Slot slots[slot_num];
char checkbits; // checkbit for this sample (optional: only in checksum mode)
} Data;
typedef struct Slot_ {
int nnz;
unsigned int* keys; // changeable to `long long` with `"input_key_type"` in `solver` object of the configuration file.
} Slot;
The Data field often has a lot of samples. Each sample starts with the labels formatted as integers and followed by nnz (number of nonzero) with the input key using the long long (or unsigned int) format as shown in Fig. 1 (a).
The input keys for categorical are distributed to the slots with no overlap allowed. For example: slot[0] = {0,10,32,45}, slot[1] = {1,2,5,67}. If there is any overlap, it will cause an undefined behavior. For example, given slot[0] = {0,10,32,45}, slot[1] = {1,10,5,67}, the table looking up the 10 key will produce different results based on how the slots are assigned to the GPUs.
File List
The first line of a file list should be the number of data files in the dataset with the paths to those files listed below as shown here:
$ cat simple_sparse_embedding_file_list.txt
10
./simple_sparse_embedding/simple_sparse_embedding0.data
./simple_sparse_embedding/simple_sparse_embedding1.data
./simple_sparse_embedding/simple_sparse_embedding2.data
./simple_sparse_embedding/simple_sparse_embedding3.data
./simple_sparse_embedding/simple_sparse_embedding4.data
./simple_sparse_embedding/simple_sparse_embedding5.data
./simple_sparse_embedding/simple_sparse_embedding6.data
./simple_sparse_embedding/simple_sparse_embedding7.data
./simple_sparse_embedding/simple_sparse_embedding8.data
./simple_sparse_embedding/simple_sparse_embedding9.data
Example:
reader = hugectr.DataReaderParams(data_reader_type = hugectr.DataReaderType_t.Norm,
source = ["./wdl_norm/file_list.txt"],
eval_source = "./wdl_norm/file_list_test.txt",
check_type = hugectr.Check_t.Sum)
Raw
The Raw dataset format is different from the Norm dataset format in that the training data appears in one binary file using int32. Fig. 1 (b) shows the structure of a Raw dataset sample.
NOTE: Only one-hot data is accepted with this format.
The Raw dataset format can be used with embedding type LocalizedSlotSparseEmbeddingOneHot only.
Example:
reader = hugectr.DataReaderParams(data_reader_type = hugectr.DataReaderType_t.Raw,
source = ["./wdl_raw/train_data.bin"],
eval_source = "./wdl_raw/validation_data.bin",
check_type = hugectr.Check_t.Sum)
Parquet
Parquet is a column-oriented, open source, and free data format. It is available to any project in the Apache Hadoop ecosystem. To reduce the file size, it supports compression and encoding. Fig. 1 © shows an example Parquet dataset. For additional information, see the parquet documentation.
Please note the following:
Nested column types are not currently supported in the Parquet data loader.
Any missing values in a column are not allowed.
Like the Norm dataset format, the label and dense feature columns should use the float format.
The Slot feature columns should use the Int64 format.
The data columns within the Parquet file can be arranged in any order.
To obtain the required information from all the rows in each parquet file and column index mapping for each label, dense (numerical), and slot (categorical) feature, a separate
_metadata.jsonfile is required.
Example _metadata.json file:
{
"file_stats":[
{
"file_name":"file0.parquet",
"num_rows":409600
},
{
"file_name":"file1.parquet",
"num_rows":409600
}
],
"cats":[
{
"col_name":"C1",
"index":4
},
{
"col_name":"C2",
"index":5
},
{
"col_name":"C3",
"index":6
},
{
"col_name":"C4",
"index":7
}
],
"conts":[
{
"col_name":"I1",
"index":1
},
{
"col_name":"I2",
"index":2
},
{
"col_name":"I3",
"index":3
}
],
"labels":[
{
"col_name":"label",
"index":0
}
]
}
reader = hugectr.DataReaderParams(data_reader_type = hugectr.DataReaderType_t.Parquet,
source = ["./parquet_data/train/_file_list.txt"],
eval_source = "./parquet_data/val/_file_list.txt",
check_type = hugectr.Check_t.Non,
slot_size_array = [10000, 50000, 20000, 300])
We provide an option to add offset for each slot by specifying slot_size_array. slot_size_array is an array whose length is equal to the number of slots. To avoid duplicate keys after adding offset, we need to ensure that the key range of the i-th slot is between 0 and slot_size_array[i]. We will do the offset in this way: for i-th slot key, we add it with offset slot_size_array[0] + slot_size_array[1] + … + slot_size_array[i - 1]. In the configuration snippet noted above, for the 0th slot, offset 0 will be added. For the 1st slot, offset 10000 will be added. And for the third slot, offset 60000 will be added. The length of slot_size_array should be equal to the length of "cats" entry in _metadata.json.
The _metadata.json is generated by NVTabular preprocessing and reside in the same folder of the file list. Basically, it contain four entries of "file_stats" (file statistics), "cats" (categorical columns), "conts" (continuous columns), and "labels" (label columns). The "col_name" and "index" in _metadata.json indicate the name and the index of a specific column in the parquet data frame. You can also edit the generated _metadata.json to only read the desired columns for model training. For example, you can modify the above _metadata.json and change the configuration correspondingly:
Example _metadata.json file after edits:
{
"file_stats":[
{
"file_name":"file0.parquet",
"num_rows":409600
},
{
"file_name":"file1.parquet",
"num_rows":409600
}
],
"cats":[
{
"col_name":"C2",
"index":5
},
{
"col_name":"C4",
"index":7
}
],
"conts":[
{
"col_name":"I1",
"index":1
},
{
"col_name":"I3",
"index":3
}
],
"labels":[
{
"col_name":"label",
"index":0
}
]
}
reader = hugectr.DataReaderParams(data_reader_type = hugectr.DataReaderType_t.Parquet,
source = ["./parquet_data/train/_file_list.txt"],
eval_source = "./parquet_data/val/_file_list.txt",
check_type = hugectr.Check_t.Non,
slot_size_array = [50000, 300])
OptParamsPy
CreateOptimizer method
hugectr.CreateOptimizer()
CreateOptimizer returns an OptParamsPy object according to the custom argument values,which specify the optimizer type and the corresponding hyperparameters. The OptParamsPy object will be used to initialize the Model instance and it applies to the weights of dense layers. Sparse embedding layers which do not have a specified optimizer will adopt this optimizer as well. Please NOTE that the hyperparameters should be configured meticulously when mixed precision training is employed, e.g., the epsilon value for the Adam optimizer should be set larger.
The embedding update supports three algorithms specified with update_type:
Local(default value): The optimizer will only update the hot columns (embedding vectors which is hit in this iteration of training) of an embedding in each iteration.Global: The optimizer will update all the columns. The embedding update type takes longer than the other embedding update types.LazyGlobal: The optimizer will only update the hot columns of an embedding in each iteration while using different semantics from the local and global updates.
Arguments
optimizer_type: The optimizer type to be used. The supported types includehugectr.Optimizer_t.Adam,hugectr.Optimizer_t.MomentumSGD,hugectr.Optimizer_t.Nesterovandhugectr.Optimizer_t.SGD,hugectr.Optimizer_t.Adagrad. The default value ishugectr.Optimizer_t.Adam.update_type: The update type for the embedding. The supported types includehugectr.Update_t.Global,hugectr.Update_t.Local, andhugectr.Update_t.LazyGlobal(Adam only). The default value ishugectr.Update_t.Global.beta1: Thebeta1value when using Adam optimizer. The default value is 0.9.beta2: Thebeta2value when using Adam optimizer. The default value is 0.999.epsilon: Theepsilonvalue when using Adam optimizer. This argument should be well configured when mixed precision training is employed. The default value is 1e-7.momentum_factor: Themomentum_factorvalue when using MomentumSGD or Nesterov optimizer. The default value is 0.atomic_update: Whether to employ atomic update when using SGD optimizer. The default value is True.
Example:
optimizer = hugectr.CreateOptimizer(optimizer_type = hugectr.Optimizer_t.Adam,
update_type = hugectr.Update_t.Global,
beta1 = 0.9,
beta2 = 0.999,
epsilon = 0.0000001)
Layers
There are three major kinds of layer in HugeCTR:
Please refer to hugectr_layer_book for detail guides on how to use different layer types.
Model
Model class
hugectr.Model()
Model groups data input, embeddings and dense network into an object with traning features. The construction of Model requires a Solver instance , a DataReaderParams instance, an OptParamsPy instance and a EmbeddingTrainingCacheParams instance (optional).
Arguments
solver: A hugectr.Solver object, the solver configuration for the model.reader_params: A hugectr.DataReaderParams object, the data reader configuration for the model.opt_params: A hugectr.OptParamsPy object, the optimizer configuration for the model.etc: A hugectr.EmbeddingTrainingCacheParams object, the embedding training cache configuration for the model. This argument should only be provided when using the embedding training cache feature.
add method
hugectr.Model.add()
The add method of Model adds an instance of Input, SparseEmbedding, DenseLayer, GroupDenseLayer, or EmbeddingCollection to the created Model object.
Typically, a Model object is comprised of one Input, several SparseEmbedding and a series of DenseLayer instances.
Please note that the loss function for HugeCTR model training is taken as a DenseLayer instance.
Arguments
inputorsparse_embeddingordense_layer: This method is an overloaded method that can accepthugectr.Input,hugectr.SparseEmbedding,hugectr.DenseLayer,hugectr.GroupDenseLayer, orhugectr.EmbeddingCollectionas an argument. It allows the users to construct their model flexibly without the JSON configuration file.
Refer to the HugeCTR Layer Classes and Methods for information about the layers and embedding collection.
compile method
hugectr.Model.compile()
This method requires no extra arguments. It allocates the internal buffer and initializes the model. For multi-task models, can optionally take two arguments.
Arguments
loss_names: List of Strings, the list of loss label names to provide weights for.loss_weights: List of Floats, the weights to be assigned to each loss label. Number of elements must match the number of loss_names.
fit method
hugectr.Model.fit()
It trains the model for a fixed number of epochs (epoch mode) or iterations (non-epoch mode). You can switch the mode of training through different configurations. To use epoch mode training, repeat_dataset within CreateSolver() should be set as False and num_epochs within Model.fit() should be set as a positive number. To use non-epoch mode training, repeat_dataset within CreateSolver() should be set as True and max_iter within Model.fit() should be set as a positive number.
Arguments
num_epochs: Integer, the number of epochs for epoch mode training. It will be ignored ifrepeat_datasetisTrue. The default value is 0.max_iter: Integer, the maximum iteration of non-epoch mode training. It will be ignored ifrepeat_datasetisFalse. The default value is 2000.display: Integer, the interval of iterations at which the training loss will be displayed. The default value is 200.eval_interval: Integer, the interval of iterations at which the evaluation will be executed. The default value is 1000.snapshot: Integer, the interval of iterations at which the snapshot model weights and optimizer states will be saved to files. This argument is invalid when embedding training cache is being used, which means no model parameters will be saved. The default value is 10000.snapshot_prefix: String, the prefix of the file names for the saved model weights and optimizer states. This argument is invalid when embedding training cache is being used, which means no model parameters will be saved. The default value is''.data_source_params: Optional, hugectr.data.DataSourceParams, a struct to specify the file system and paths to use while dumping the models.
summary method
hugectr.Model.summary()
This method takes no extra arguments and prints a string summary of the model. Users can have an overview of the model structure with this method.
graph_to_json method
hugectr.Model.graph_to_json()
This method saves the model graph to a JSON file, which can be used for continuous training and inference.
Arguments
graph_config_file: The JSON file to which the model graph will be saved. There is NO default value and it should be specified by users.
construct_from_json method
hugectr.Model.construct_from_json()
This method constructs the model graph from a saved JSON file, which is useful for continuous training and fine-tune.
Arguments
graph_config_file: The saved JSON file from which the model graph will be constructed. There is NO default value and it should be specified by users.include_dense_network: Boolean, whether to include the dense network when constructing the model graph. If it isTrue, the whole model graph will be constructed, then both saved sparse model weights and dense model weights can be loaded. If it isFalse, only the sparse embedding layers will be constructed and the corresponding sparse model weights can be loaded, which enables users to construct a new dense network on top of that. Please NOTE that the HugeCTR layers are organized by names and you can check the input name, output name and output shape and of the added layers withModel.summary(). There is NO default value and it should be specified by users.
load_dense_weights method
hugectr.Model.load_dense_weights()
This method load the dense weights from the saved dense model file.
Arguments
dense_model_file: String, the saved dense model file from which the dense weights will be loaded. There is NO default value and it should be specified by users.data_source_params: Optional, hugectr.data.DataSourceParams, a struct to specify the file system and paths to use while loading the dense model. Ifdata_source_params.use_hdfsis set toFalse,dense_model_filewill be used as the path.
load_dense_optimizer_states method
hugectr.Model.load_dense_optimizer_states()
This method load the dense optimizer states from the saved dense optimizer states file.
Arguments
dense_opt_states_file: String, the saved dense optimizer states file from which the dense optimizer states will be loaded. There is NO default value and it should be specified by users.data_source_params: Optional, hugectr.data.DataSourceParams, a struct to specify the file system and paths to use while loading the dense optimizer states. Ifdata_source_params.use_hdfsis set toFalse,dense_opt_states_filewill be used as the path.
load_sparse_weights method
hugectr.Model.load_sparse_weights()
This method load the sparse weights from the saved sparse embedding files.
Implementation Ⅰ
Arguments
sparse_embedding_files: List[str], the sparse embedding files from which the sparse weights will be loaded. The number of files should equal to that of the sparse embedding layers in the model. There is NO default value and it should be specified by users.data_source_params: Optional, hugectr.data.DataSourceParams, a struct to specify the file system and paths to use while loading the sparse models. Ifdata_source_params.use_hdfsis set toFalse,sparse_embedding_fileswill be used as the path.
Implementation Ⅱ
Arguments
sparse_embedding_files_map: Dict[str, str], the sparse embedding file will be loaded by the embedding layer with the specified sparse embedding name. There is NO default value and it should be specified by users.
Example:
model.add(hugectr.SparseEmbedding(embedding_type = hugectr.Embedding_t.DistributedSlotSparseEmbeddingHash,
workspace_size_per_gpu_in_mb = 23,
embedding_vec_size = 1,
combiner = "sum",
sparse_embedding_name = "sparse_embedding2",
bottom_name = "wide_data",
optimizer = optimizer))
model.add(hugectr.SparseEmbedding(embedding_type = hugectr.Embedding_t.DistributedSlotSparseEmbeddingHash,
workspace_size_per_gpu_in_mb = 358,
embedding_vec_size = 16,
combiner = "sum",
sparse_embedding_name = "sparse_embedding1",
bottom_name = "deep_data",
optimizer = optimizer))
# ...
model.load_sparse_weights(["wdl_0_sparse_4000.model", "wdl_1_sparse_4000.model"]) # load models for both embedding layers
model.load_sparse_weights({"sparse_embedding1": "wdl_1_sparse_4000.model"}) # or load the model for one embedding layer
load_sparse_optimizer_states method
hugectr.Model.load_sparse_optimizer_states()
This method load the sparse optimizer states from the saved sparse optimizer states files.
Implementation Ⅰ
Arguments
sparse_opt_states_files: List[str], the sparse optimizer states files from which the sparse optimizer states will be loaded. The number of files should equal to that of the sparse embedding layers in the model. There is NO default value and it should be specified by users.data_source_params: Optional, hugectr.data.DataSourceParams, a struct to specify the file system and paths to use while loading the sparse optimizer states. Ifdata_source_params.use_hdfsis set toFalse,sparse_opt_states_fileswill be used as the path.
Implementation Ⅱ
Arguments
sparse_opt_states_files_map: Dict[str, str], the sparse optimizer states file will be loaded by the embedding layer with the specified sparse embedding name. There is NO default value and it should be specified by users.
freeze_dense method
hugectr.Model.freeze_dense()
This method takes no extra arguments and freezes the dense weights of the model. Users can use this method when they want to fine-tune the sparse weights.
freeze_embedding method
hugectr.Model.freeze_embedding()
Implementation Ⅰ: freeze the weights of all the embedding layers. This method takes no extra arguments and freezes the sparse weights of the model. Users can use this method when they only want to train the dense weights.
Implementation Ⅱ: freeze the weights of a specific embedding layer. Please refer to Section 3.4 of HugeCTR Criteo Notebook for the usage.
Arguments
embedding_name: String, the name of the embedding layer.
Example:
model.add(hugectr.SparseEmbedding(embedding_type = hugectr.Embedding_t.DistributedSlotSparseEmbeddingHash,
workspace_size_per_gpu_in_mb = 23,
embedding_vec_size = 1,
combiner = "sum",
sparse_embedding_name = "sparse_embedding2",
bottom_name = "wide_data",
optimizer = optimizer))
model.add(hugectr.SparseEmbedding(embedding_type = hugectr.Embedding_t.DistributedSlotSparseEmbeddingHash,
workspace_size_per_gpu_in_mb = 358,
embedding_vec_size = 16,
combiner = "sum",
sparse_embedding_name = "sparse_embedding1",
bottom_name = "deep_data",
optimizer = optimizer))
# ...
model.freeze_embedding() # freeze all the embedding layers
model.freeze_embedding("sparse_embedding1") # or free a specific embedding layer
unfreeze_dense method
hugectr.Model.unfreeze_dense()
This method takes no extra arguments and unfreezes the dense weights of the model.
unfreeze_embedding method
hugectr.Model.unfreeze_embedding()
Implementation Ⅰ: unfreeze the weights of all the embedding layers. This method takes no extra arguments and unfreezes the sparse weights of the model.
Implementation Ⅱ: unfreeze the weights of a specific embedding layer.
Arguments
embedding_name: String, the name of the embedding layer.
reset_learning_rate_scheduler method
hugectr.Model.reset_learning_rate_scheduler()
This method resets the learning rate scheduler of the model. Users can use this method when they want to fine-tune the model weights.
Arguments
base_lr: The base learning rate for the internal learning rate scheduler within Model instance. There is NO default value and it should be specified by users.warmup_steps: The warmup steps for the internal learning rate scheduler within Model instance. The default value is 1.decay_start: The step at which the learning rate decay starts for the internal learning rate scheduler within Model instance. The default value is 0.decay_steps: The number of steps of the learning rate decay for the internal learning rate scheduler within Model instance. The default value is 1.decay_power: The power of the learning rate decay for the internal learning rate scheduler within Model instance. The default value is 2.end_lr: The final learning rate for the internal learning rate scheduler within Model instance. The default value is 0.
set_source method
hugectr.Model.set_source()
The set_source method can set the data source and keyset files under epoch mode training. This overloaded method has two implementations.
Implementation Ⅰ: only valid when repeat_dataset is False and use_embedding_training_cache is True.
Arguments
source: List[str], the training dataset source. It can be specified with several file lists, e.g.,source = ["file_list.1.txt", "file_list.2.txt"]. There is NO default value and it should be specified by users.keyset: List[str], the keyset files. It should be corresponding to thesource. For example, we can specifysource = ["file_list.1.txt", "file_list.2.txt"]andsource = ["file_list.1.keyset", "file_list.2.keyset"], which have a one-to-one correspondence. There is NO default value and it should be specified by users.eval_source: String, the evaluation dataset source. There is NO default value and it should be specified by users.
Implementation Ⅱ: only valid when repeat_dataset is False and use_embedding_training_cache is False.
Arguments
source: String, the training dataset source. For Norm or Parquet dataset, it should be the file list of training data. For Raw dataset, it should be a single training file. There is NO default value and it should be specified by users.eval_source: String, the evaluation dataset source. For Norm or Parquet dataset, it should be the file list of evaluation data. For Raw dataset, it should be a single evaluation file. There is NO default value and it should be specified by users.
Low-level Training API
For HugeCTR low-level training API, the core data structures are basically the same as the high-level training API. On this basis, we expose the internal LearningRateScheduler, DataReader and EmbeddingTrainingCache within the Model, and provide some low-level training methods as well.HugeCTR currently supports both epoch mode training and non-epoch mode training for dataset in Norm and Raw formats, and only supports non-epoch mode training for dataset in Parquet format. While introducing the API usage, we will elaborate how to employ these two modes of training.
LearningRateScheduler
get_next method
hugectr.LearningRateScheduler.get_next()
This method takes no extra arguments and returns the learning rate to be used for the next iteration.
DataReader
set_source method
hugectr.DataReader32.set_source()
hugectr.DataReader64.set_source()
The set_source method of DataReader currently supports the dataset in Norm and Raw formats, and should be used in epoch mode training. When the data reader reaches the end of file for the current training data or evaluation data, this method can be used to re-specify the training data file or evaluation data file.
Arguments
file_name: The file name of the new training source or evaluation source. For Norm format dataset, it takes the form offile_list.txt. For Raw format dataset, it appears asdata.bin. The default value is'', which means that the data reader will reset to the beginning of the current data file.
is_eof method
hugectr.DataReader32.is_eof()
hugectr.DataReader64.is_eof()
This method takes no extra arguments and returns whether the data reader has reached the end of the current source file.
EmbeddingTraingCache
update method
hugectr.EmbeddingTraingCache.update()
The update method of EmbeddingTraingCache currently supports Norm format datasets. Using this method requires that a series of file lists and the corresponding keyset files are generated at the same time when preprocessing the original data to Norm format. This method gives you the ability to load a subset of an embedding table into the GPU in a coarse grained, on-demand manner during the training stage. Please refer to HugeCTR Embedding Traing Cache if you want to get detailed information about EmbeddingTraingCache.
Arguments
keyset_fileorkeyset_file_list: This method is an overloaded method that can accept str or List[str] as an argument. For the model with multiple embedding tables, if the keyset of each embedding table is not separated when generating the keyset files, then pass in thekeyset_file. If the keyset of each embedding table has been separated when generating keyset files, you need to pass in thekeyset_file_list, the size of which should equal to the number of embedding tables.
Model
get_learning_rate_scheduler method
hugectr.Model.get_learning_rate_scheduler()
hugectr.Model.get_learning_rate_scheduler generates and returns the LearningRateScheduler object of the model instance. When the SGD optimizer is adopted for training, the returned object can obtain the dynamically changing learning rate according to the warmup_steps, decay_start and decay_steps configured in the hugectr.CreateSolver method.
Refer to SGD Optimizer and Learning Rate Scheduling) if you want to get detailed information about LearningRateScheduler.
get_embedding_training_cache method
hugectr.Model.get_embedding_training_cache()
This method takes no extra arguments and returns the EmbeddingTrainingCache object.
get_data_reader_train method
hugectr.Model.get_data_reader_train()
This method takes no extra arguments and returns the DataReader object that reads the training data.
get_data_reader_eval method
hugectr.Model.get_data_reader_eval()
This method takes no extra arguments and returns the DataReader object that reads the evaluation data.
start_data_reading method
hugectr.Model.start_data_reading()
This method takes no extra arguments and should be used if and only if it is under the non-epoch mode training. The method starts the train_data_reader and eval_data_reader before entering the training loop.
set_learning_rate method
hugectr.Model.set_learning_rate()
This method is used together with the get_next method of LearningRateScheduler and sets the learning rate for the next training iteration.
Arguments
lr: Float, the learning rate to be set。
train method
hugectr.Model.train()
This method takes no extra arguments and executes one iteration of the model weights based on one minibatch of training data.
get_current_loss method
hugectr.Model.get_current_loss()
This method takes no extra arguments and returns the loss value for the current iteration.
eval method
hugectr.Model.eval()
This method takes no arguments and calculates the evaluation metrics based on one minibatch of evaluation data.
get_eval_metrics method
hugectr.Model.get_eval_metrics()
This method takes no extra arguments and returns the average evaluation metrics of several minibatches of evaluation data.
get_incremental_model method
updated_model = hugectr.Model.get_incremental_model()
This method is only supported in Embedding Training Cache and returns the updated embedding table since the last time calling this method to updated_model. Note that updated_model only stores the embedding features being touched instead of the whole table.
When training with multi-node, the updated_model returned in each node doesn’t have duplicated embedding features, and the aggregations of updated_model from each node form the complete updated sparse model.
The length of updated_model is equal to the number of embedding tables in your model, e.g., length(updated_model)==2 for the wdl model. Each element in updated_model is a pair of NumPy arrays: a 1-D array stores keys in long long format, and another 2-D array stores embedding vectors in float format, where the leading dimension is the embedding vector size. E.g., updated_model[0][0] stores keys, and updated_model[0][1] stores the embedding vectors corresponding to keys in updated_model[0][0].
dump_incremental_model_2kafka method
hugectr.Model.dump_incremental_model_2kafka()
This method is only supported in Embedding Training Cache. It with post the updated embedding table to Kafka as user specified.
Please NOTE that is method can not be used together with the get_incremental_model method. Only one of these two methods could be used for dumping the incremental model.
save_params_to_files method
hugectr.Model.save_params_to_files()
This method save the model parameters to files. If Embedding Training Cache is utilized, this method will save sparse weights, dense weights and dense optimizer states. Otherwise, this method will save sparse weights, sparse optimizer states, dense weights and dense optimizer states.
The stored sparse model can be used for both the later training and inference cases. Each sparse model will be dumped as a separate folder that contains two files (key, emb_vector) for the DistributedSlotEmbedding or three files (key, slot_id, emb_vector) for the LocalizedSlotEmbedding. Details of these files are:
key: The unique keys appeared in the training data. All keys are stored inlong longformat, and HugeCTR will handle the datatype conversion internally for the case wheni64_input_key = False.slot_id: The key distribution info internally used by the LocalizedSlotEmbedding.emb_vector: The embedding vectors corresponding to keys stored in thekeyfile.
Note that the key, slot id, and embedding vector are stored in the sparse model in the same sequence, so both the nth slot id in slot_id file and the nth embedding vector in the emb_vector file are mapped to the nth key in the key file.
Arguments
prefix: String, the prefix of the saved files for model weights and optimizer states. There is NO default value and it should be specified by users.iter: Integer, the current number of iterations, which will be the suffix of the saved files for model weights and optimizer states. The default value is 0.
export_predictions method
hugectr.Model.export_predictions()
If you want to export the predictions for specified data, using predict() in inference API is recommended. This method will export the last batch of evaluation prediction and label to file. If the file already exists, the evaluation result will be appended to the end of the file. This method will only export eval_batch_size evaluation result each time. So it should be used in the following way:
for i in range(train_steps):
# do train
...
# clean prediction / label result file
prediction_file_in_current_step = "predictions" + str(i)
if os.path.exists(prediction_file_in_current_step):
os.remove(prediction_file_in_current_step)
label_file_in_current_step = "label" + str(i)
if os.path.exists(label_file_in_current_step):
os.remove(label_file_in_current_step)
# do evaluation and export prediction
for _ in range(solver.max_eval_batches):
model.eval()
model.export_predictions(prediction_file_in_current_step, label_file_in_current_step)
Arguments
output_prediction_file_name: String, the file to which the evaluation prediction results will be written. The order of the prediction results are the same as that of the labels, but may be different with the order of the samples in the dataset. There is NO default value and it should be specified by users.output_label_file_name: String, the file to which the evaluation labels will be written. The order of the labels are the same as that of the prediction results, but may be different with the order of the samples in the dataset. There is NO default value and it should be specified by users.
Inference API
For HugeCTR inference API, the core data structures are InferenceParams and InferenceModel. They are designed and implemented for the purpose of multi-GPU offline inference. Please refer to HugeCTR Backend if online inference with Triton is needed.
Please NOTE that Inference API requires a configuration JSON file of the model graph, which can derived from the Model.graph_to_json() method. Besides, model_name within CreateSolver should be specified during training in order to correctly dump the JSON file.
InferenceParams
InferenceParams class
hugectr.inference.InferenceParams()
InferenceParams specifies the parameters related to the inference. An InferenceParams instance is required to initialize the InferenceModel instance.
Refer to the HPS Configuration documentation for the parameters.
InferenceModel
InferenceModel class
hugectr.inference.InferenceModel()
InferenceModel is a collection of inference sessions deployed on multiple GPUs, which can leverage Hierarchical Parameter Server and enable concurrent execution. The construction of InferenceModel requires a model configuration file and an InferenceParams instance.
Arguments
model_config_path: String, the inference model configuration file (which can be derived fromModel.graph_to_json). There is NO default value and it should be specified by users.inference_params: InferenceParams, the InferenceParams object. There is NO default value and it should be specified by users.
predict method
hugectr.inference.InferenceModel.predict()
The predict method of InferenceModel makes predictions based on the dataset of Norm or Parquet format. It will return the 2-D numpy array of the shape (max_batchsize*num_batches, label_dim), whose order is consistent with the sample order in the dataset. If max_batchsize*num_batches is greater than the total number of samples in the dataset, it will loop over the dataset. For example, there are totally 40000 samples in the dataset, max_batchsize equals 4096, num_batches equals 10 and label_dim equals 2. The returned array will be of the shape (40960, 2), of which first 40000 rows should be desired results and the last 960 rows correspond to the first 960 samples in the dataset.
Arguments
num_batches: Integer, the number of prediction batches.source: String, the source of prediction dataset. It should be the file list for Norm or Parquet format data.data_reader_type:hugectr.DataReaderType_t, the data reader type. We supporthugectr.DataReaderType_t.Normandhugectr.DataReaderType_t.Parquet.check_type:hugectr.Check_t, the check type for the data source. We currently supporthugectr.Check_t.Sumandhugectr.Check_t.Non.slot_size_array: List[int], the cardinality array of input features. It should be consistent with that of the sparse input. We requires this argument for Parquet format data. The default value is an empty list, which is suitable for Norm format data.
evaluate method
hugectr.inference.InferenceModel.evaluate()
The evaluate method of InferenceModel does evaluations based on the dataset of Norm or Parquet format. It requires that the dataset contains the label field. This method returns the AUC value for the specified evaluation batches.
Arguments
num_batches: Integer, the number of evaluation batches.source: String, the source of evaluation dataset. It should be the file list for Norm or Parquet format data.data_reader_type:hugectr.DataReaderType_t, the data reader type. We supporthugectr.DataReaderType_t.Normandhugectr.DataReaderType_t.Parquet.check_type:hugectr.Check_t, the check type for the data source. We supporthugectr.Check_t.Sumandhugectr.Check_t.Noncurrently.slot_size_array: List[int], the cardinality array of input features. It should be consistent with that of the sparse input. We requires this argument for Parquet format data. The default value is an empty list, which is suitable for Norm format data.
Data Generator API
For HugeCTR data generator API, the core data structures are DataGeneratorParams and DataGenerator. Please refer to data_generator directory in the HugeCTR repository on GitHub to acknowledge how to write Python scripts to generate synthetic dataset and start training HugeCTR model.
DataGeneratorParams class
hugectr.tools.DataGeneratorParams()
DataGeneratorParams specifies the parameters related to the data generation. An DataGeneratorParams instance is required to initialize the DataGenerator instance.
Arguments
format: The format for synthetic dataset. The supported types includehugectr.DataReaderType_t.Norm,hugectr.DataReaderType_t.Parquetandhugectr.DataReaderType_t.Raw. There is NO default value and it should be specified by users.label_dim: Integer, the label dimension for synthetic dataset. There is NO default value and it should be specified by users.dense_dim: Integer, the number of dense (or continuous) features for synthetic dataset. There is NO default value and it should be specified by users.num_slot: Integer, the number of sparse feature slots for synthetic dataset. There is NO default value and it should be specified by users.i64_input_key: Boolean, whether to use I64 for input keys for synthetic dataset. If your dataset format is Norm or Paruqet, you can choose the data type of each input key. For the Raw dataset format, only I32 is allowed. There is NO default value and it should be specified by users.source: String, the synthetic training dataset source. For Norm or Parquet dataset, it should be the file list of training data, e.g., source = “file_list.txt”. For Raw dataset, it should be a single training file, e.g., source = “train_data.bin”. There is NO default value and it should be specified by users.eval_source: String, the synthetic evaluation dataset source. For Norm or Parquet dataset, it should be the file list of evaluation data, e.g., source = “file_list_test.txt”. For Raw dataset, it should be a single evaluation file, e.g., source = “test_data.bin”. There is NO default value and it should be specified by users.slot_size_array: List[int], the cardinality array of input features for synthetic dataset. The list length should be equal tonum_slot. There is NO default value and it should be specified by users.nnz_array: List[int], the number of non-zero entries in each slot for synthetic dataset. The list length should be equal tonum_slot. This argument helps to simulate one-hot or multi-hot encodings. The default value is an empty list and one-hot encoding will be employed then.check_type: The data error detection mechanism. The supported types includehugectr.Check_t.Sum(CheckSum) andhugectr.Check_t.Non(no detection). The default value ishugectr.Check_t.Sum.dist_type: The distribution of the sparse input keys for synthetic dataset. The supported types includehugectr.Distribution_t.PowerLawandhugectr.Distribution_t.Uniform. The default value ishugectr.Distribution_t.PowerLaw.power_law_type: The specific distribution of power law distribution. The supported types includehugectr.PowerLaw_t.Long(alpha=0.9),hugectr.PowerLaw_t.Medium(alpha=1.1),hugectr.PowerLaw_t.Short(alpha=1.3) andhugectr.PowerLaw_t.Specific(requiring a specific alpha value). This argument is only valid whendist_typeishugectr.Distribution_t.PowerLaw. The default value ishugectr.PowerLaw_t.Specific.alpha: Float, the alpha value for power law distribution. This argument is only valid whendist_typeishugectr.Distribution_t.PowerLawandpower_law_typeishugectr.PowerLaw_t.Specific. The alpha value should be greater than zero and not equal to 1.0. The default value is 1.2.num_files: Integer, the number of training data files that will be generated. This argument is valid whenformatishugectr.DataReaderType_t.Normorhugectr.DataReaderType_t.Parquet. The default value is 128.eval_num_files: Integer, the number of evaluation data files that will be generated. This argument is valid whenformatishugectr.DataReaderType_t.Normorhugectr.DataReaderType_t.Parquet. The default value is 32.num_samples_per_file: Integer, the number of samples per generated data file. This argument is valid whenformatishugectr.DataReaderType_t.Normorhugectr.DataReaderType_t.Parquet. The default value is 40960.num_samples: Integer, the number of samples in the generated single training data file (e.g., train_data.bin). This argument is only valid whenformatishugectr.DataReaderType_t.Raw. The default value is 5242880.eval_num_samples: Integer, the number of samples in the generated single evaluation data file (e.g., test_data.bin). This argument is only valid whenformatishugectr.DataReaderType_t.Raw. The default value is 1310720.float_label_dense: Boolean, this is only valid whenformatishugectr.DataReaderType_t.Raw. If its value is set to True, the label and dense features for each sample are interpreted as float values. Otherwise, they are regarded as integer values while the dense features are preprocessed with log(dense[i] + 1.f). The default value is False.
DataGenerator
DataGenerator class
hugectr.tools.DataGenerator()
DataGenerator provides an API to generate synthetic Norm, Parquet or Raw dataset. The construction of DataGenerator requires a DataGeneratorParams instance.
Arguments
data_generator_params: The DataGeneratorParams instance which encapsulates the required parameters for data generation. There is NO default value and it should be specified by users.
generate method
hugectr.tools.DataGenerator.generate()
This method takes no extra arguments and starts to generate the synthetic dataset based on the configurations within data_generator_params.
Data Source API
The data source API is used to specify which file system to use in the following training process. There are two data structures: DataSourceParams and DataSource. Please refer to data_source directory in the HugeCTRrepository on GitHub to check out how to write Python scripts to move data from HDFS to local FS.
DataSourceParams class
hugectr.data.DataSourceParams()
DataSourceParams specifies the file system information and the paths to data and model used for training. A DataSourceParams instance is required to initialize the DataSource instance.
Arguments
source:hugect.DataSourceType_t, can beLocalorHDFS, specifying the file system. Default ishugectr.DataSourceType_t.Local.server: String, the IP address of your file system. For Hadoop cluster, it is your namenode. Will be ignored ifsourceisDataSourceType_t.Local. Default is ‘localhost’.port: Integer, the port to listen from your server. Will be ignored ifsourceisDataSourceType_t.Local. Default is 9000.